Visualizing how pathogens spread and change
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202303_VizBi.html
Human seasonal influenza viruses

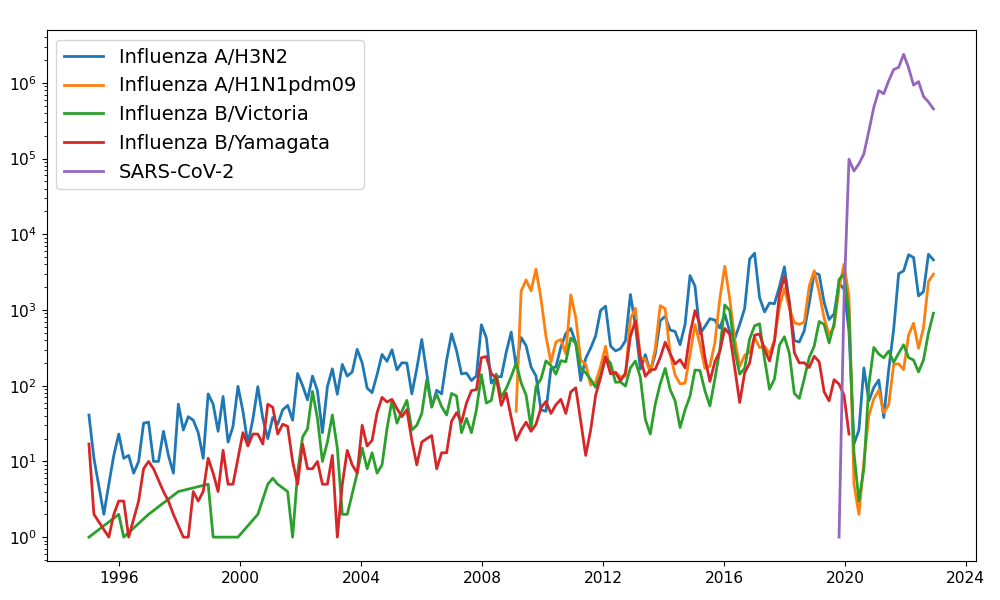
Positive tests for influenza in the USA by week
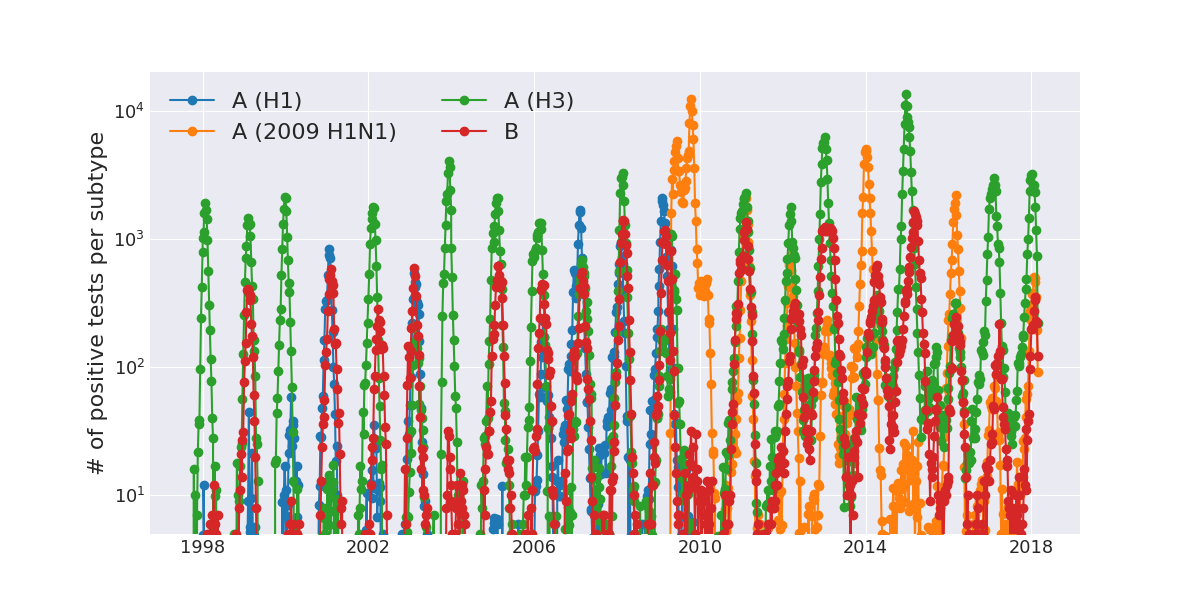 Data by the US CDC
Data by the US CDC
Virus genomes change rapidly through time
A/Brisbane/100/2014
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... hundreds of thousands of sequences...
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... hundreds of thousands of sequences...
Genome sequences record the spread of pathogens




in SARS-CoV-2 a new mutation accumulates about every 2 weeks



- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

Many data dimensions
- Phylogenetic tree -- hierarchical relatedness structure
- Genotype -- mutations and variants
- Dynamics and time
- Spatial distribution
- Phenotypes
Nextstrain.org
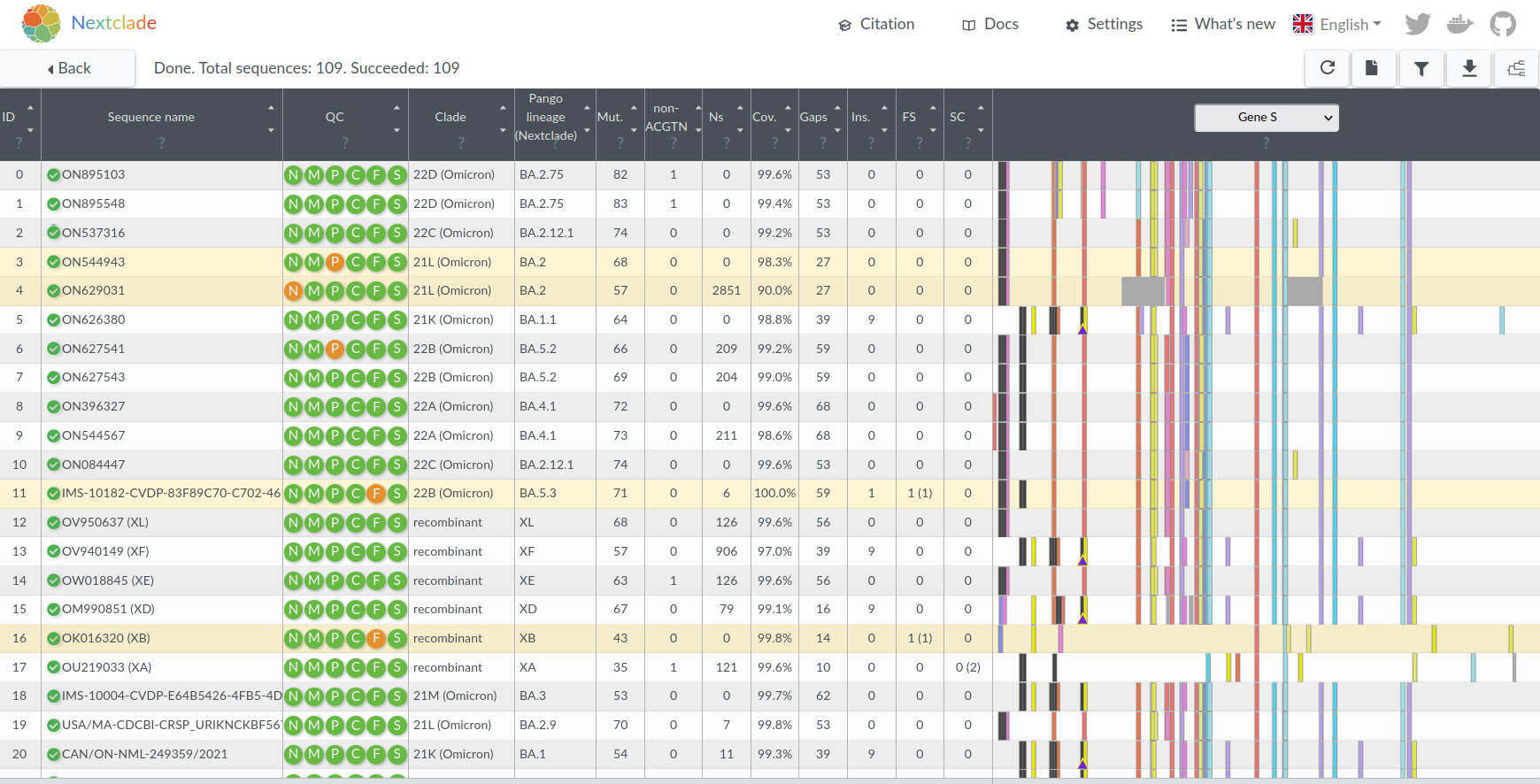
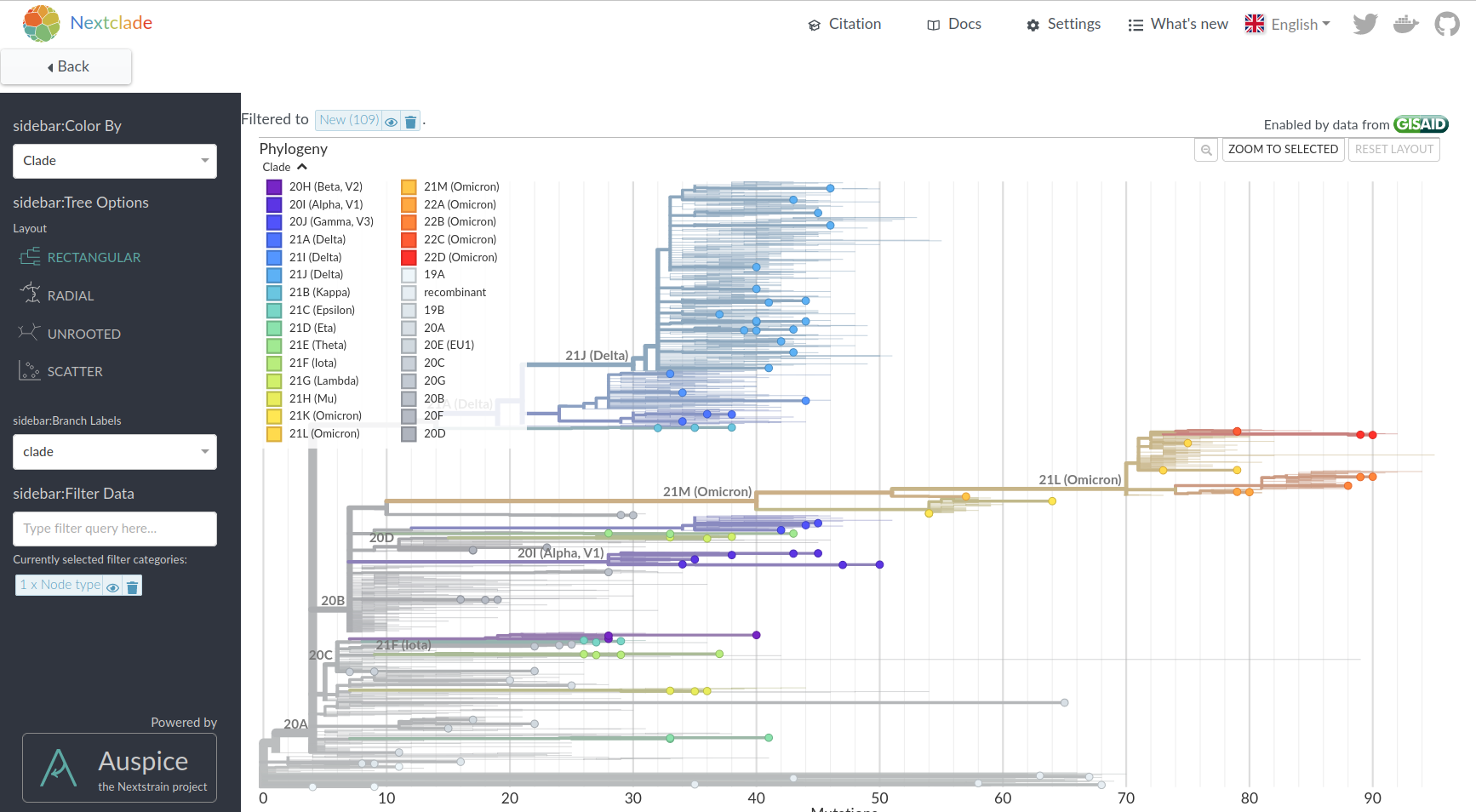
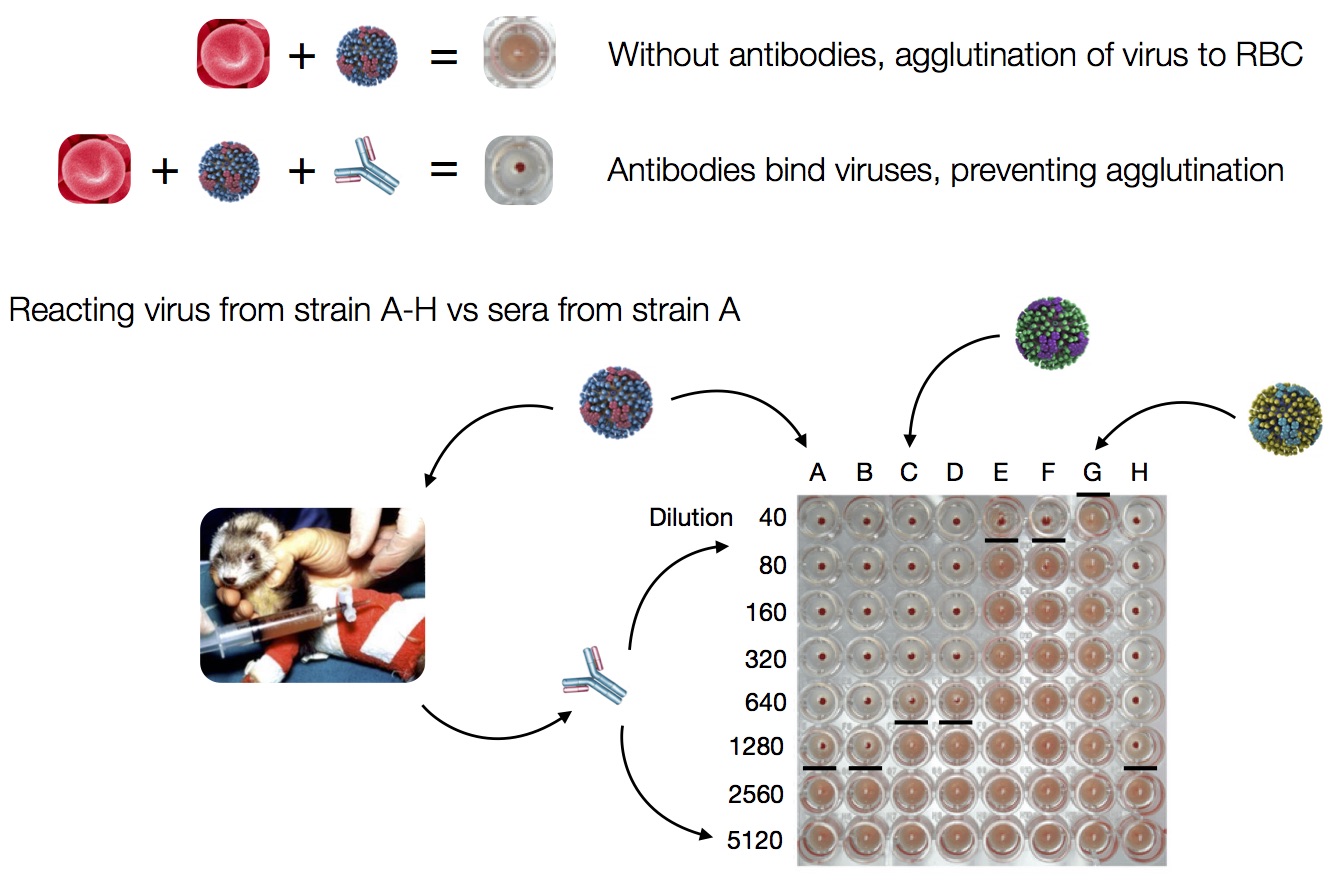
Hemagglutination Inhibition assays
Antigenic distance tables
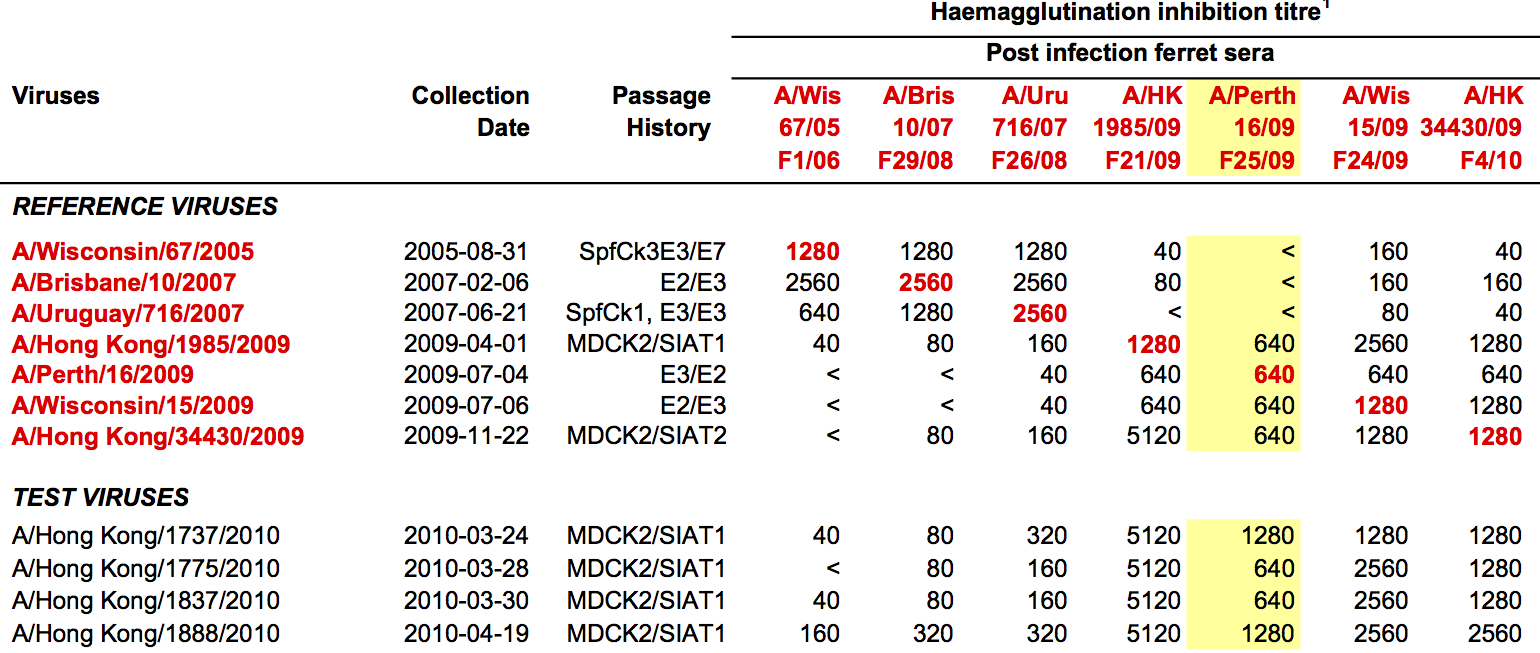
- Long list of distances between sera and viruses
- Tables are sparse, only close by pairs
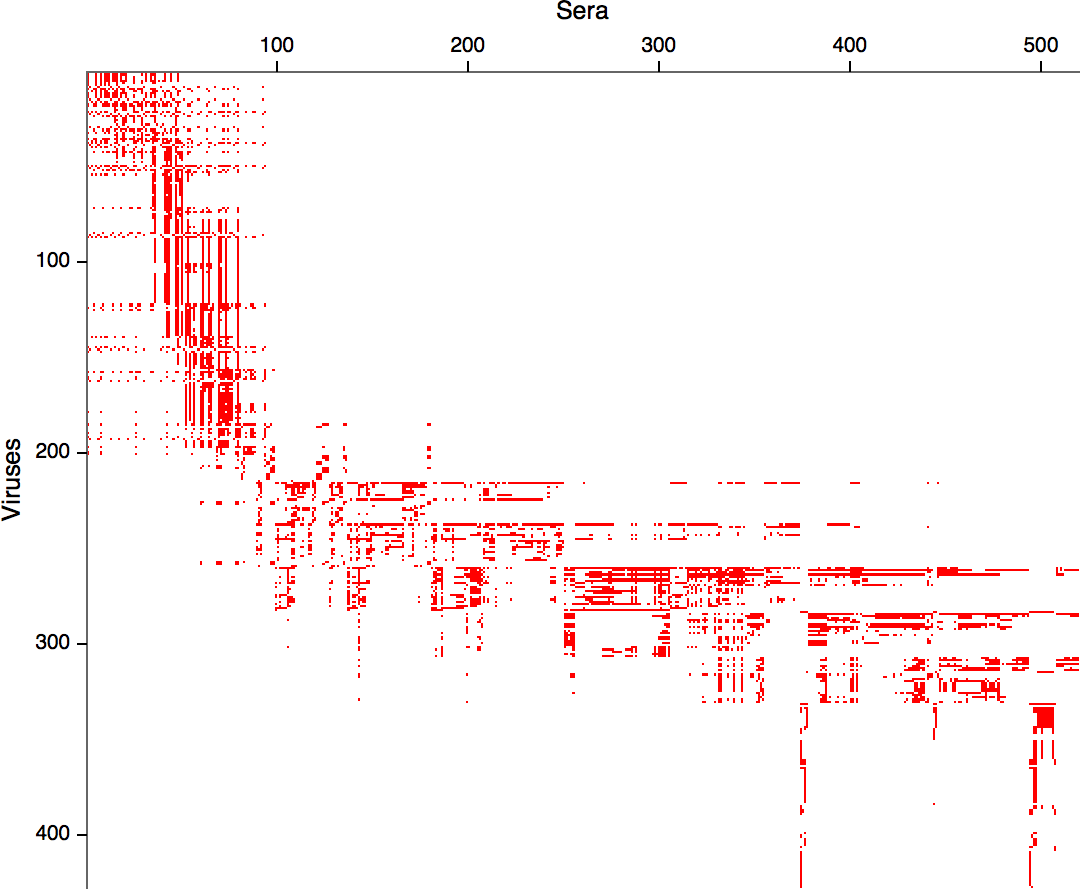
Integrating antigenic and molecular evolution
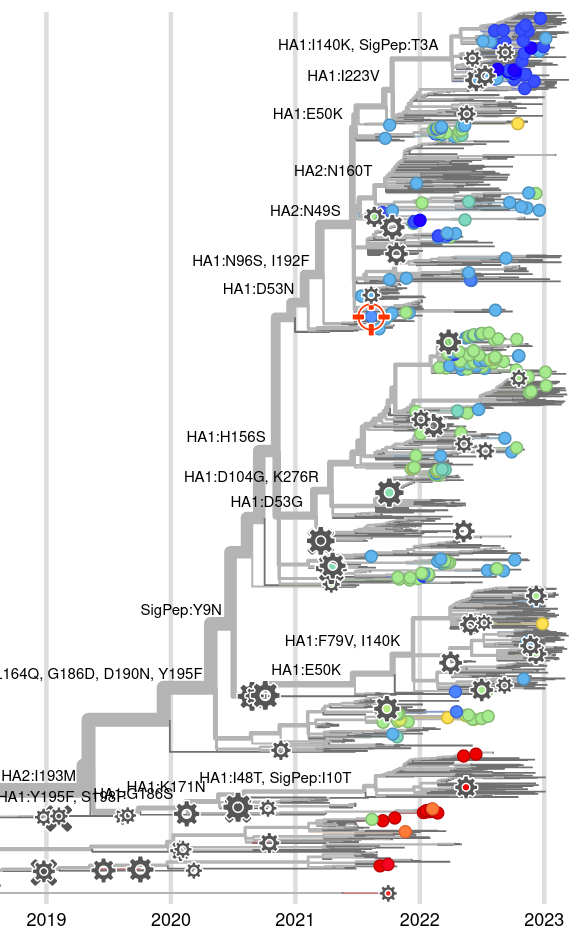
 RN et al, PNAS, 2017
RN et al, PNAS, 2017
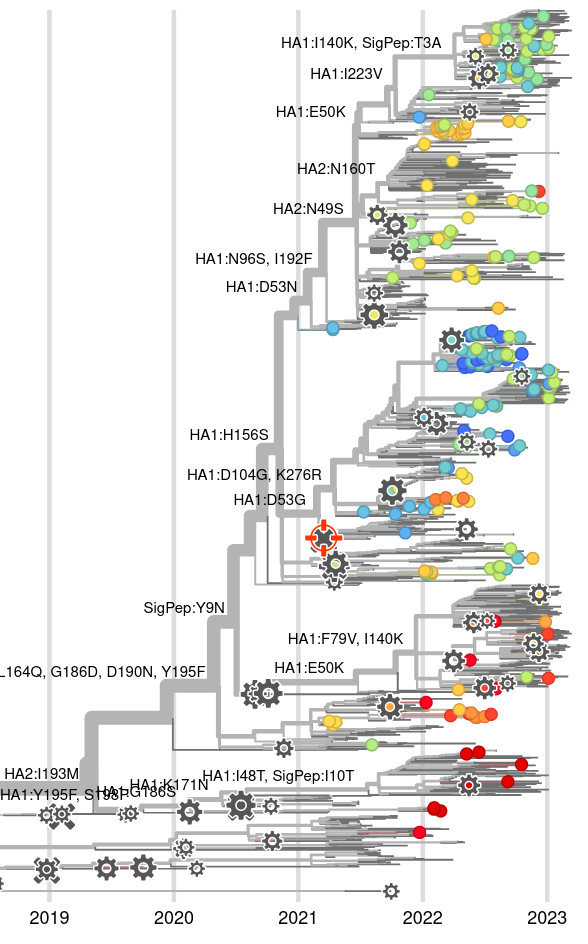
Integrating antigenic and molecular evolution
 RN et al, PNAS, 2017
RN et al, PNAS, 2017
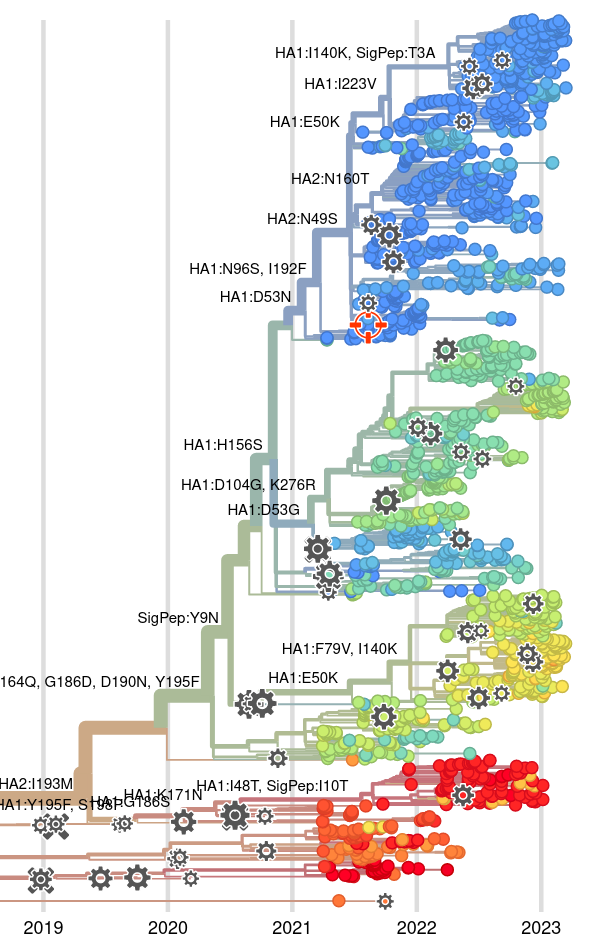
Integrating antigenic and molecular evolution
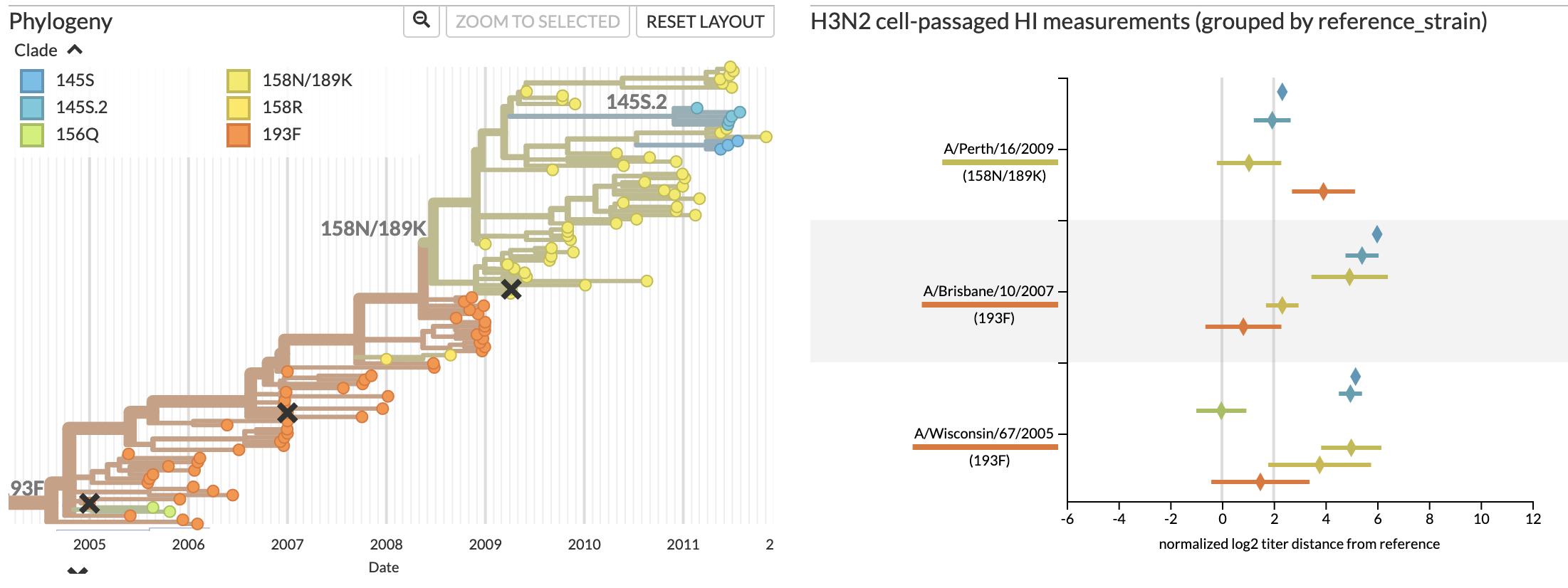 Jover Lee..., John Huddleston, 2023
Jover Lee..., John Huddleston, 2023
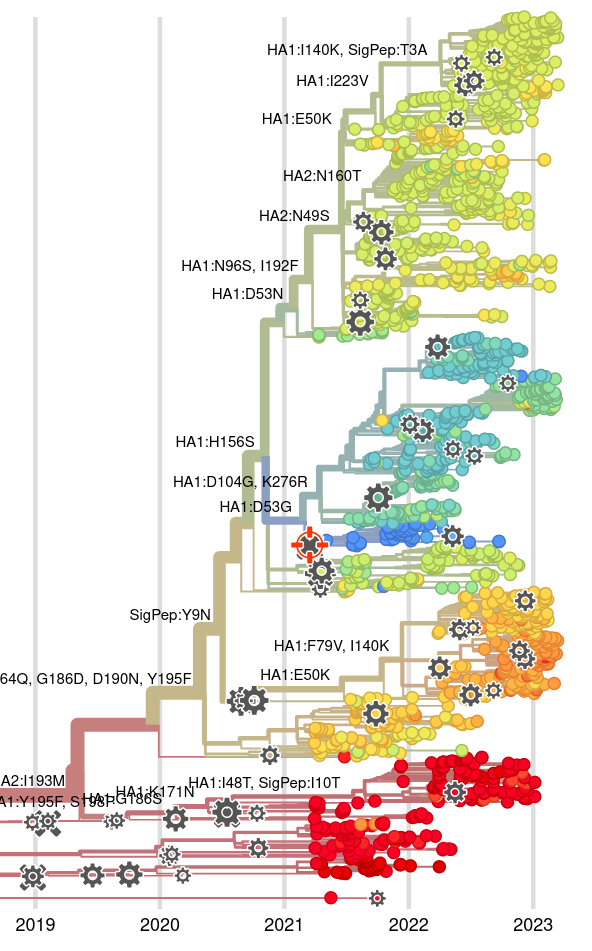
Integrating antigenic and molecular evolution
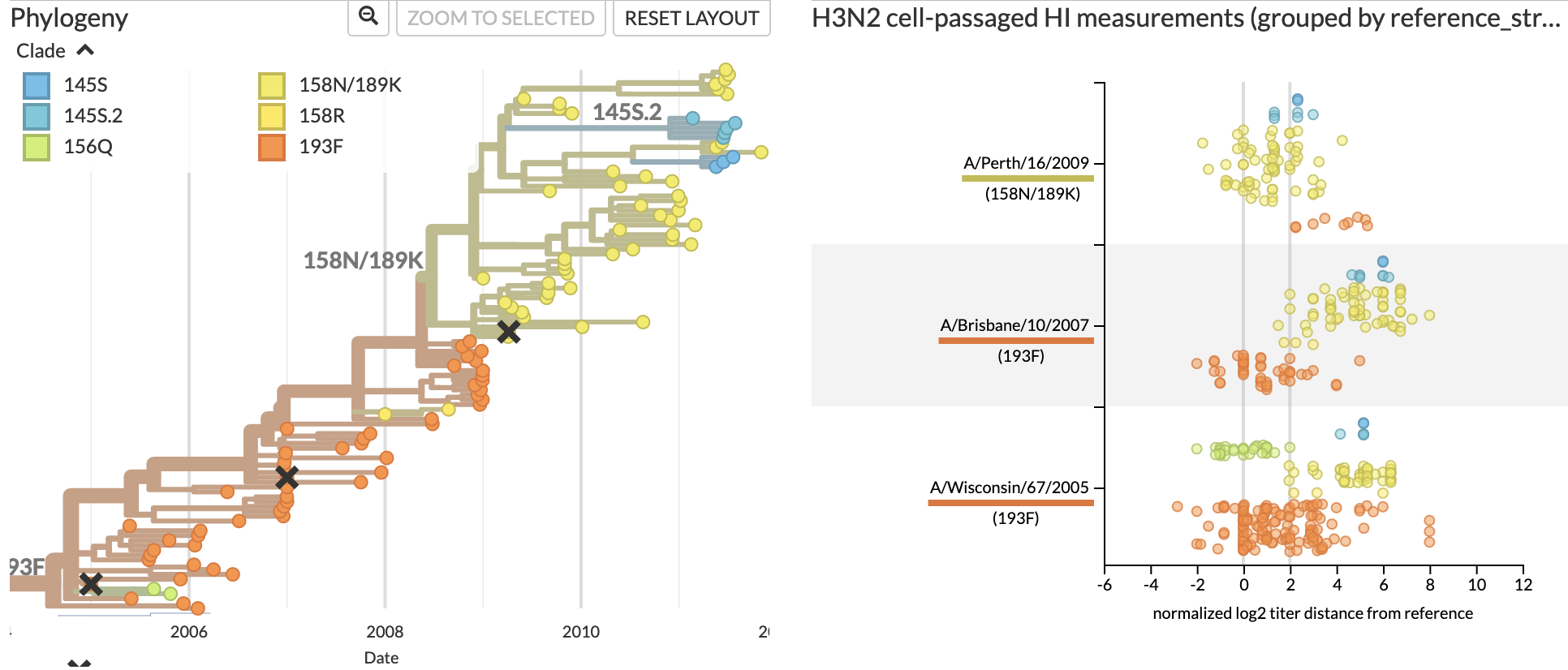 Jover Lee..., John Huddleston, 2023
Jover Lee..., John Huddleston, 2023
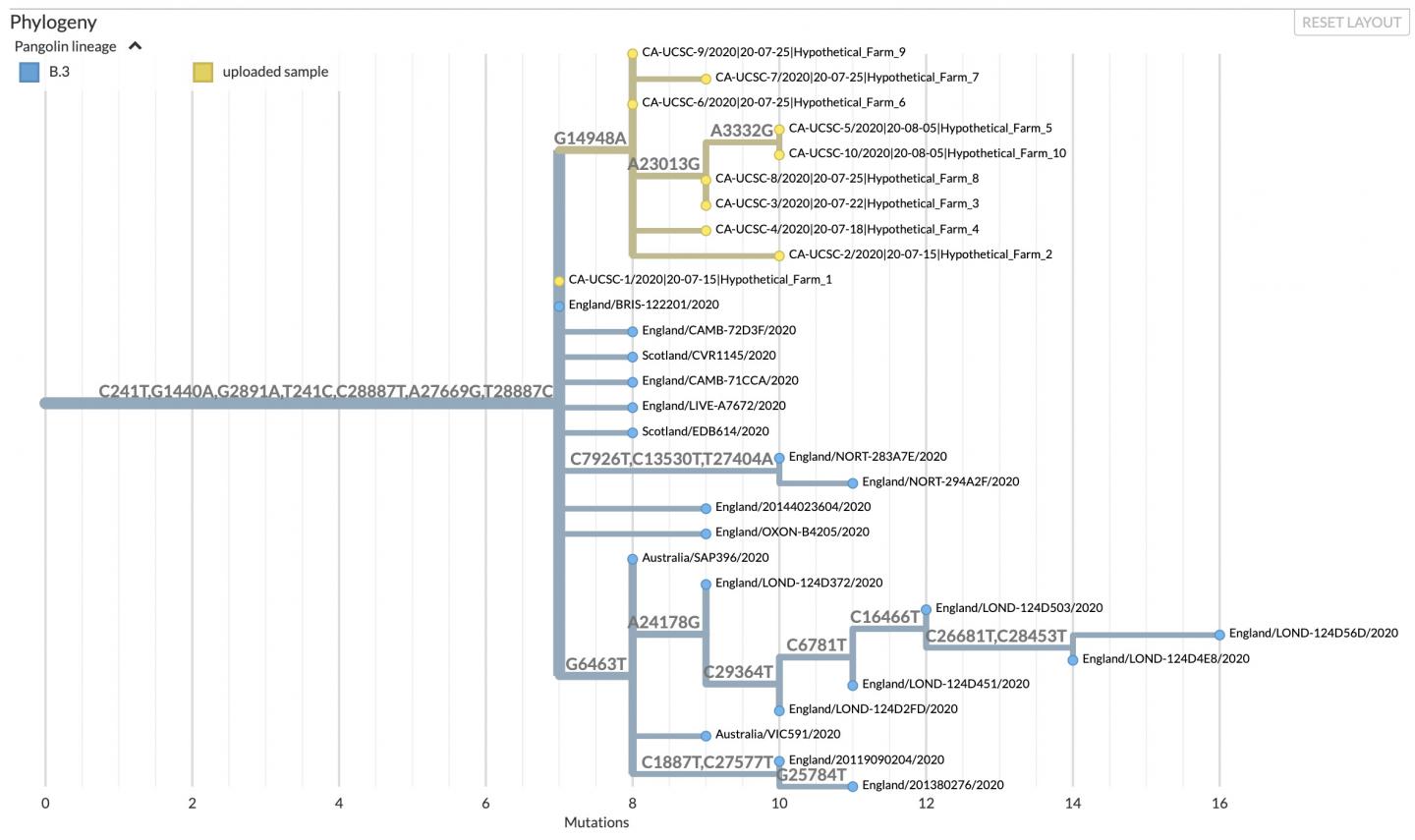
None of this works for millions of tips
Taxonium by Theo Sanderson
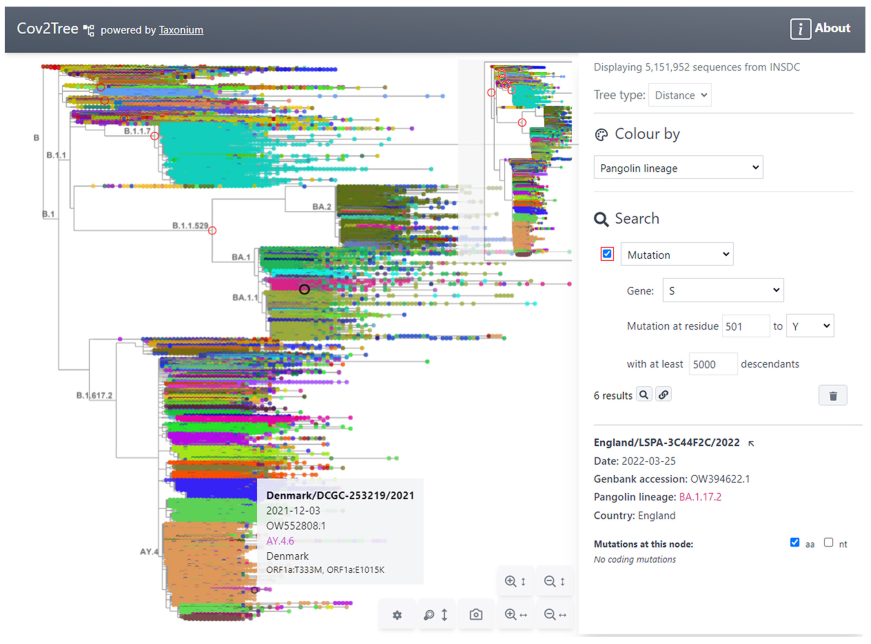
Pandemic scale trees by UShER
- phylogenetic tree of millions of samples grown through placement
- Fast search on this tree to explore the context of specific samples
- Extraction of the context and visualization in Nextstrain
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Cornelius Roemer, Moira Zuber, and John Huddleston
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID

