In a collaboration with the Sommer lab, we have a new paper out in Genetics. We report whole genome resequencing of 104 globally sampled strains of Pristionchus pacificus and the draft genome of a close outgroup P. expectatus. The strains have been collected and sequenced by the group of Ralf Sommer here at the MPI in Tuebingen. This represents one of the largest data sets to analyze population genetic processes in a natural population.
Evolution and population genetics on multiple time scales
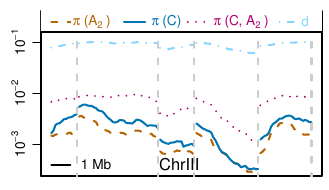
The population is diverse and highly structured, which allows us to
study evolution at various time scales. Intra-clade diversity is about
0.3% and inter-clade diversity about 1%, while the outgroup is roughly
10% diverged. Strong population structure indicates that there is little
shared variation across clades, but the chromosomal diversity profiles
(see figure) are strongly correlated across clades suggesting that
similar, but independent,
 {.wp-image-264
.alignleft width="240"
height="138"}
processes shape genetic diversity. Together with the observation that an
increasing amount of non-synonymous variation is lost with increasing
separation and a negative correlation of local gene density and
diversity suggests purifying selection as dominant force shaping
diversity. LD, however, extends over many megabases and the dynamics of
most polymorphisms is predominantly determined by its genetic background
rather than its own effect on fitness. The latter only manifests itself
on time scales comparable to the separation of clades.
{.wp-image-264
.alignleft width="240"
height="138"}
processes shape genetic diversity. Together with the observation that an
increasing amount of non-synonymous variation is lost with increasing
separation and a negative correlation of local gene density and
diversity suggests purifying selection as dominant force shaping
diversity. LD, however, extends over many megabases and the dynamics of
most polymorphisms is predominantly determined by its genetic background
rather than its own effect on fitness. The latter only manifests itself
on time scales comparable to the separation of clades.
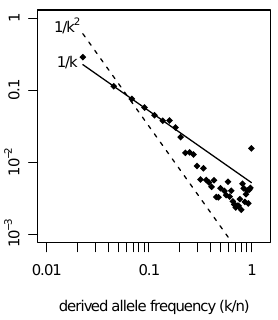
Site frequency spectra are shaped by drift and draft
Linked selection acting on many loci -- whether deleterious or
beneficial does not matter -- results in distortions of genealogies with
characteristic effects on the site frequency spectrum (SFS). In
particular, one expects a U-shaped spectrum with an excess of rare and
high frequency derived alleles. After careful polarization, this is
exactly what we
see.
 {.wp-image-263
.alignright width="208"
height="240"}
Up to around frequencies of 10%, the SFS is compatible with neutral models
(1/k decay), but at intermediate frequencies it resembles the SFS
expected in presence of linked selection. This is plausible, as there is
ample opportunity for linked selection: most non-synonymous
polymorphisms are selected against and LD extends over several
megabases. While individual effects might be weak, the collective
aggregation of long linked haplotypes can exert strong selection.
{.wp-image-263
.alignright width="208"
height="240"}
Up to around frequencies of 10%, the SFS is compatible with neutral models
(1/k decay), but at intermediate frequencies it resembles the SFS
expected in presence of linked selection. This is plausible, as there is
ample opportunity for linked selection: most non-synonymous
polymorphisms are selected against and LD extends over several
megabases. While individual effects might be weak, the collective
aggregation of long linked haplotypes can exert strong selection.
Why do we care?
Neutral models have long been used to interpret genetic diversity data -- but they rarely fit well, at least when there is enough data. Rather than forcing the data into a particular model framework, we have attempted to find informative observables. The U-shaped SFS, for example, clearly show the inadequacies of neutral neutral models. The SFS suggest that drift-like effects are important on short time scales, but that genetic diversity is shaped by different processes on longer time scales. Those processes probably have little to do with the population size and caution is warranted when using inference methods based on neutral null models. While linked selection on predominantly deleterious variation is a plausible cause for these non-drift like features, further research is necessary to tell to what extent ecological processes are shaping genetic diversity in this species.
Christian Rodelsperger and Richard Neher