Nextstrain consists of two components
Augur is composed of a series of modules and different workflows will use different parts of the pipeline. A selection of augur modules and different possible entry points are illustrated below.
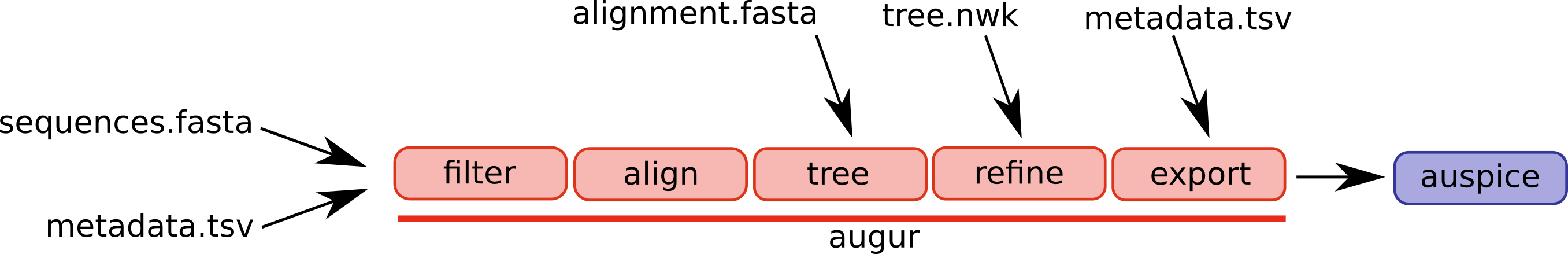
The canonical pipeline would ingest sequences and metadata such as dates and sampling locations, filter the data, align the sequences, infer a tree, and export the results in a format that can be visualized by auspice. For a step by step explanation on how your analysis is built up using augur steps, see this slide deck.
In some cases, you might start with a manually curated alignment and want to start the workflow at the tree building step.
Or you already have a tree inferred. In this case, you only need to feed you tree through the refine and export steps.
The refine step is necessary to ensure that cross-referencing between tree nodes and meta data works as expected.
For a full list of augur modules, see the augur documentation.
Augur includes for example modules for phylogeographic analysis and ancestral sequences reconstruction.
The different augur modules can be strung together by workflow managers like snakemake and nextflow. The nextstrain team uses snakemake to run and manage the different analysis that you see on nextstrain.org.