We are thrilled and humbled that nextstrain.org was selected as the winner of the OpenSciencePrize. The OpenSciencePrize is an initiative from the National Institutes of Health (US), the Wellcome Trust (UK), and the Howard Hughes Medical Institute. The prize was awarded in a three state competition. In the first stage, six out of about 100 teams were selected to develop a prototype and present their results in Washington DC in December 2016. All of these teams presented exciting contributions -- I personally liked OpenAQ or MyGene2 a lot. The public got to select three out of these six finalists and an expert panel chose nextstrain.org among those three.
NextStrain would not be possible without open sharing of sequence data and we are indebted to all data producers -- in particular Nick Loman, Paris Sabeti, Kristian Andersen, Andrew Rambaut, the Sanger Center and many others for early sharing and being forceful advocates of open data. We hope that nextstrain.org will help fighting viral diseases more effective, advance our understanding of how viral pathogens evolve, and provide incentives to openly share viral sequence data.
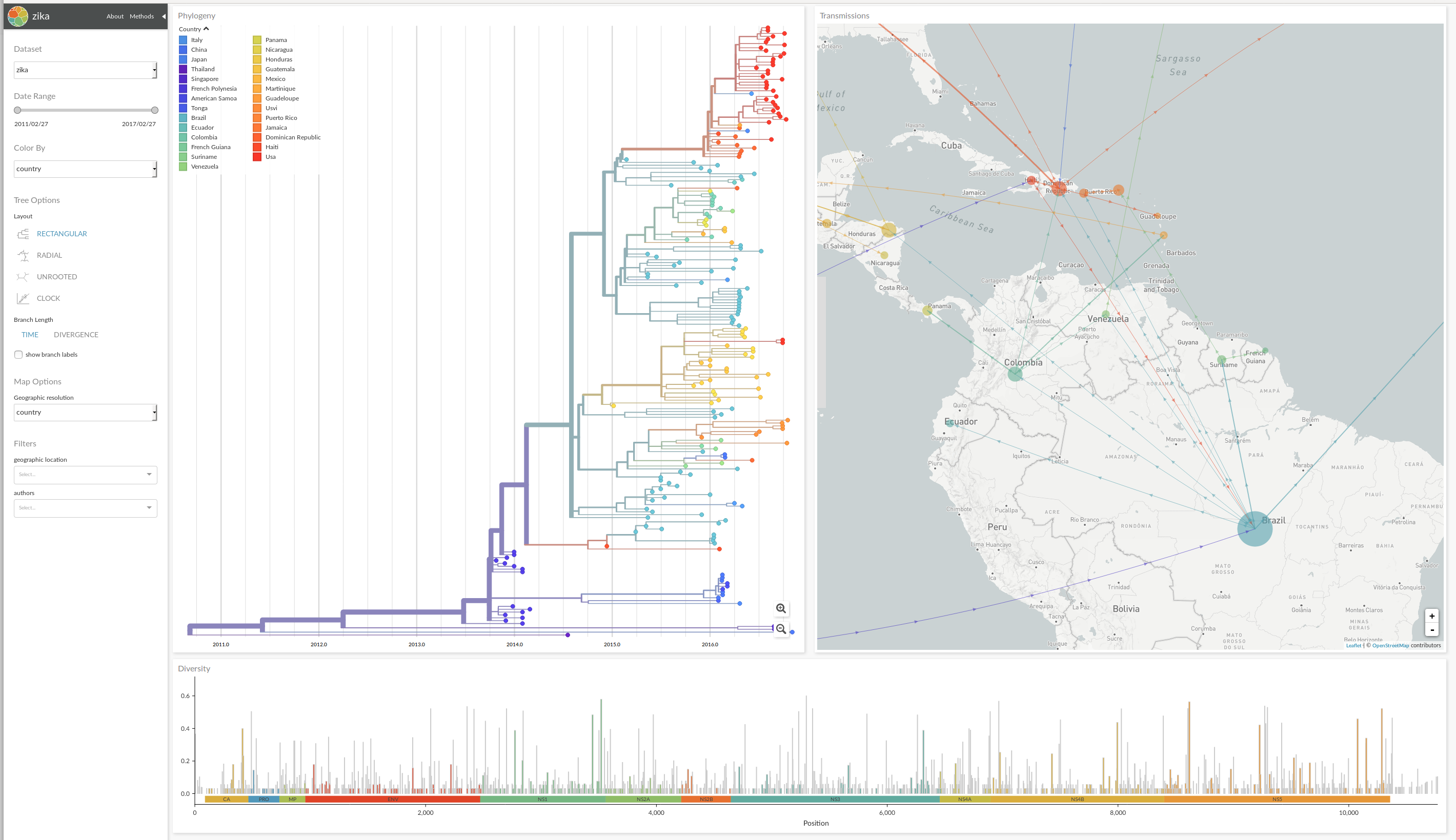
nextstrain.org grew out of Trevor's and my efforts to track and predict the evolution of influenza viruses at nextflu.org. The code base of nextflu has grown organically and has become pretty messy as we kept adding new features. About a year ago, we decided to do a complete rewrite and consult outside expertise. Having been selected as winners of the OpenSciencePrize has pushed us to release nextstrain 2.0 along with the announcement of the prize. nextstrain.org now features a multipanel layout including a map and multiple different tree views.
While nextflu and nextstrain 1.0 was cobbled together by Trevor and me, nextstrain 2.0 was a true team effort to which many people contributed: Colin Megill put together much of the front-end and taught as react.js, Pavel Sagulenko developed treetime which we use to rapidly calculate time trees, Charlton Callender has developed a data base to manage the sequence and meta data, and James Hadfield joint us recently but has already put in a huge amount of work to make the front-end work.
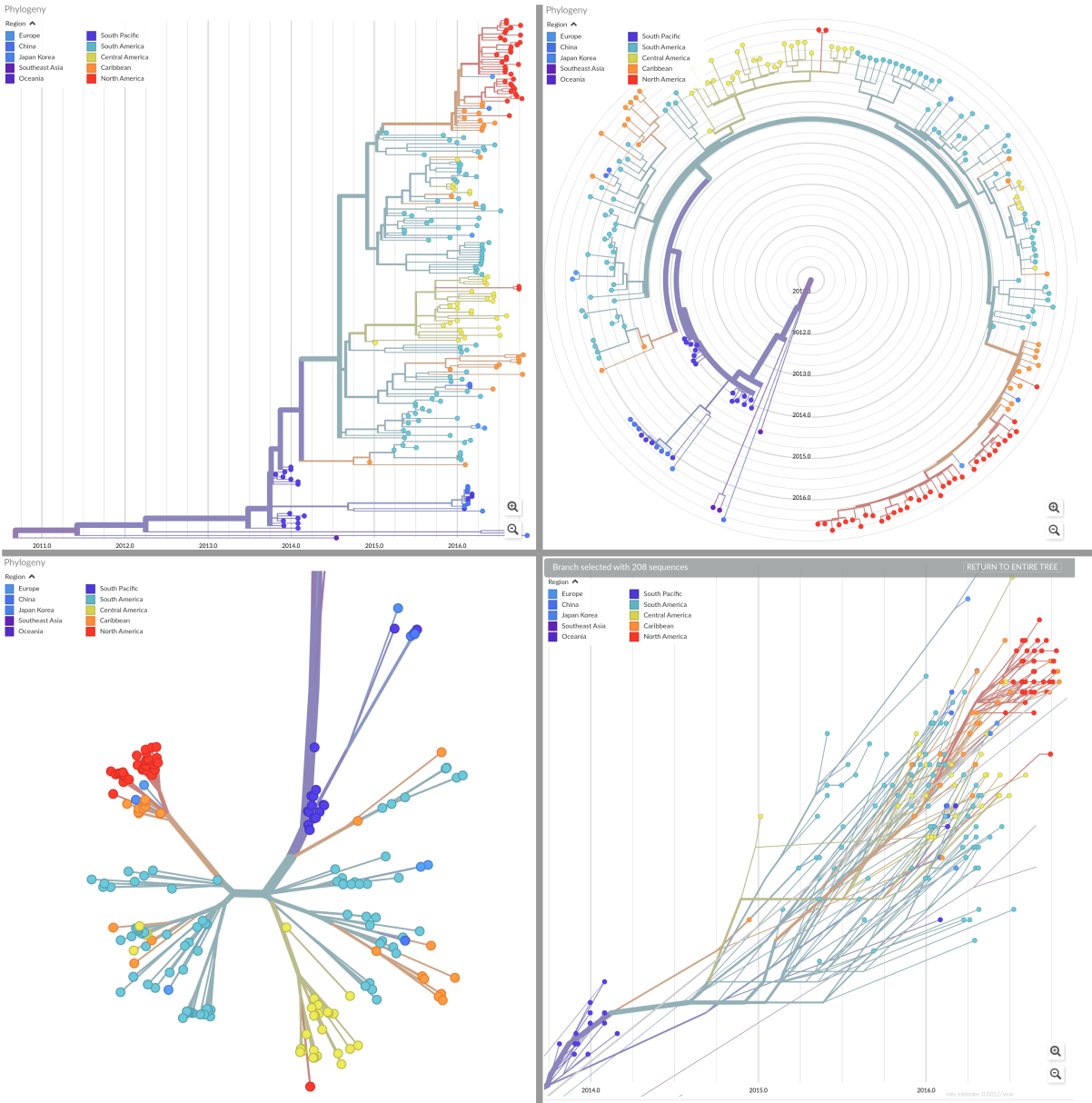
The most significant innovation compared to version v1 is a map which now shows where on the globe cases have been along with plausible path along which the viruses have traveled. But also the phylogenetic tree viewer got a bunch of new features. The tree can be zoomed and panned and informative tooltips provide meta data and context. You can toggle between a timetree and a regular divergence tree and choose between different tree layouts. The rectangular, radial, unrooted, and "clock" layout are shown below. The "clock" layout plots the sampling time vs sequence divergence and is useful to assess the rate of evolution of a virus.
Please play around at nextstrain.org and let us know if you have any comments, suggestions, or questions. We still have a number of things that we want to accomplish soon, for example a tighter integration of the map with the tree, extension to more viruses, etc.