The wide spread use of antibiotic has lead to evolution and spread of bacteria resistant to drugs. Multi-resistant strains tend to circulate in hospitals with a high density of people on antibiotic therapy. Treatment options are running out for some of the most resistant strains. To limit the spread and emergence of such variants, we need to understand the conditions under which resistance evolves, and how we can avoid or delay it.
Matthias Willmann and Silke Peter from the Tübingen medical school and my group teamed up to study evolution of colistin resistance in clinical isolates of Pseudomonas aeruginosa using a morbidostat. This computer controlled continuous culture device was designed by Erdal Toprak and colleagues in Roy Kishony's lab. The morbidostat adjust drug concentrations such that the bugs struggle but grow. Over days and weeks, the bacteria become resistant while we can take samples and sequence the population to track the changes in their genomes.
The preprint we just posted describes results by Bianca Regenbogen who took on the ambitious task of i) finalizing our morbidostat set-up, (ii) establishing the experimental procedures, (iii) running the experiments for two strains (PA77 and PA83), (iv) sequencing the sample, and (v) analysing the data. Every part of this would have been impressive for a six month thesis - she did all five.
The morbidostat
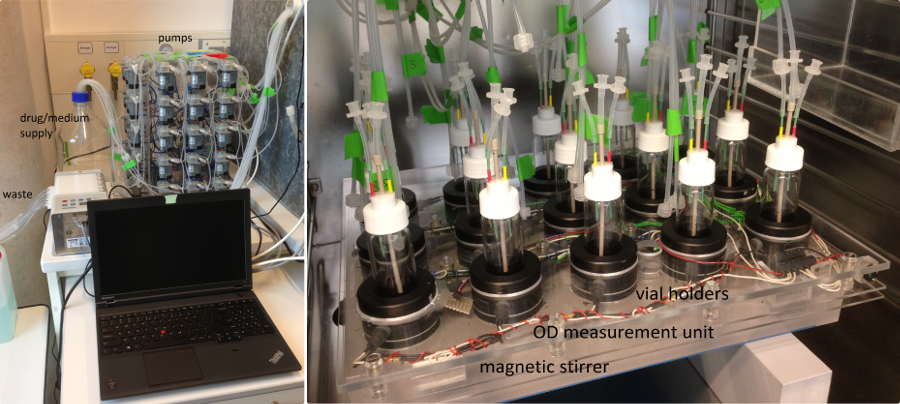
The left part of the picture shows the laptop controlling the experiment and a large array of small pumps that deliver drug and growth medium into the culture vials. The culture vials sit inside an incubator (right picture). All the tubing connecting pumps to the vials and for waste removal run through a hole that our workshop drilled into the back of incubator (yes, they did ask whether we were serious about this).
The experiments
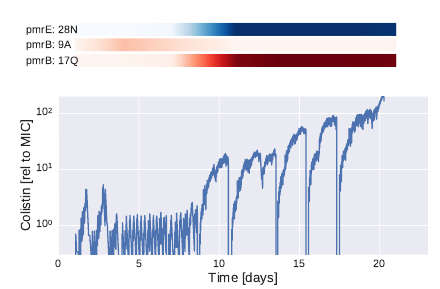
The experiments typically ran for 3 weeks, while we recorded optical density every 30s, adjusted medium and drug every 10min, and took samples three times per week. The plot on the left shows the colistin concentration over these three weeks - we have a total of 18 such traces, each corresponding to one culture. The colistin concentrations tolerated by the bacteria increased 10fold after 7-12 days, and then steadily increased further to a total of a 100fold increase. The colored bars above the graph indicated the trajectories of commonly observed mutations: more below.
The mutations
We wanted to know what happened as these populations became resistant.
Which genes mutated? Always the same genes? Same mutations? .... We
sequenced the seven samples for each culture (\~100 samples) with high
coverage. In one vial, the dynamics of mutations looked like this:
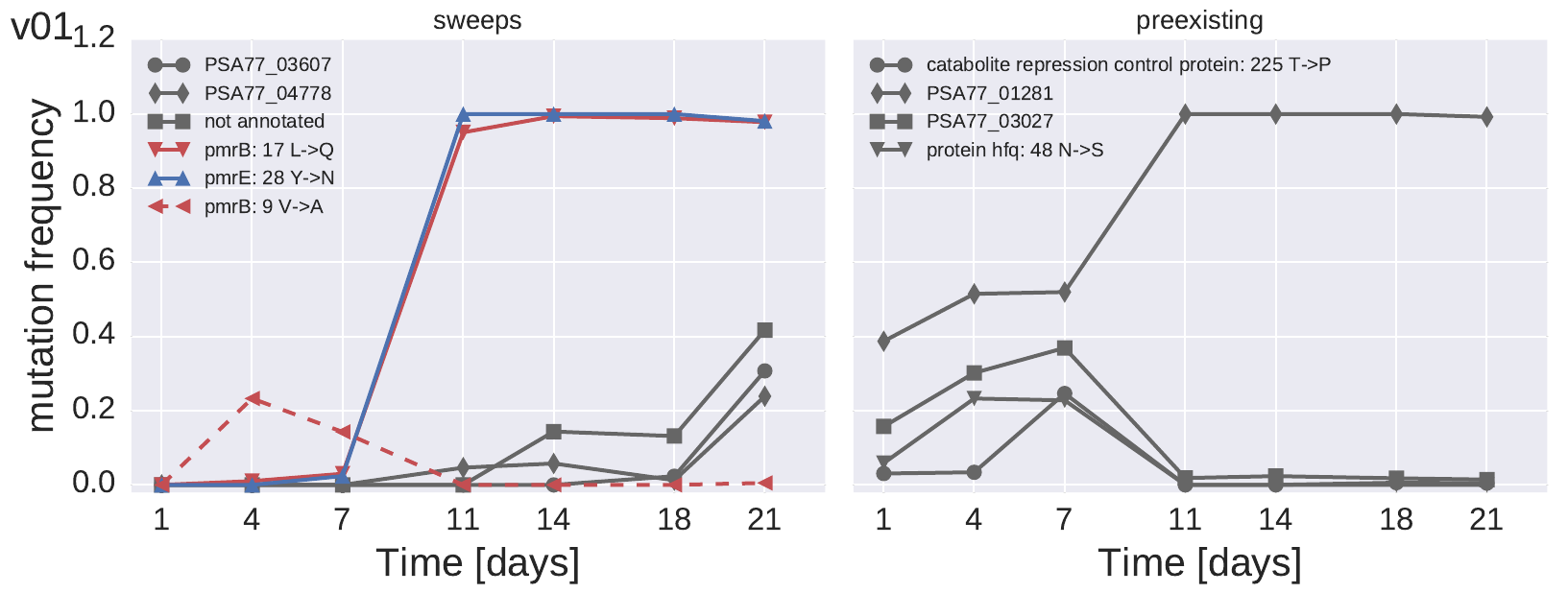
The left panel shows the trajectories of mutations that we didn't see in initial sample, the right panel are the preexisting mutations. The plot only shows mutations observed at >20% at least once.
All PA77 cultures mutated pmrE and pmrB. All mutations in pmrE were at position 28, either to Y->C or Y->N. In contrast, mutations in pmrB were all over the place. In the case shown above, two pmrB mutations came up, but only L17Q together with Y28N in pmrE was successful. Curiously, PA77 is an outlier at position 28 of pmrE: The majority of P. aeruginosa strains in GenBank have a C at this position.
PA83 evolved colistin resistance via much more varied patterns of mutations. While we didn't find any mutations in pmrE, all strains mutated lpxC, pmrB, 7/9 mutated migA (+2 other poorly annotated genes), 6/9 lpxO2, 5/9 pmrA. In total more than 20 different genes mutated in more than 2 cultures.
The above mutation seem to be directly related to colistin resistance and most mutated genes are involved in some part of the synthesis of lipopolysaccharides in the outer membrane. In addition to those directly relevant mutations, 6/9 and 1/9 cultures of PA83 and PA77, respectively, carried mutations in mutS. These populations had a 100fold elevated mutation rate and developed many other unique mutations (\~50 in 3 weeks).
What did we learn?
- a rich culture of 20ml (100M cells) develops resistance in 10 days
- the pathways differ between strains: LpxC, migA,.. mutations were common in PA83, not observed in PA77 PmrE reverted to wildtype in PA77, PA83 has pmrE 28C
- despite different pathways, phenotypes evolve in a similar fashion
- mutator phenotype arise frequently, but are not essential for resistance evolution
- the mutation target for colistin is large and clones with similar mutations compete.
- MICs measurements in liquid culture differ from those of ETests.