
related posts
Bacteria are incredibly diverse and inhabit almost every accessible place on this planet. They are essential symbionts of plants and animals, but also formidable pathogens. Most of our work so far has been theoretical or on evolution of viruses and we only recently got into bacterial evolution. In one project, we have performed experimental evolution to study the emergence of drug resistance. In another project, we are developing tools to analyze and visualize bacterial pan-genomes.
Drug resistance evolution in a morbidostat
Pathogenic bacteria are increasingly resistant against many antibiotics we use to treat infections and resistance is developing into a serious complication of modern medicine. Matthias Willmann and Silke Peter from the Tübingen medical school and our group teamed up to study evolution of colistin resistance in clinical isolates of Pseudomonas aeruginosa using a morbidostat. This computer controlled continuous culture device was designed by Erdal Toprak and colleagues in Roy Kishony's lab. The morbidostat adjust drug concentrations such that the bugs struggle but grow. Over days and weeks, the bacteria become resistant while we can take samples and sequence the population to track the changes in their genomes. Over 2-3 weeks, all replicate cultures became resistant via mutational pathways that are reproducible, but strain dependent (Regenbogen et al, AAC, 2017).
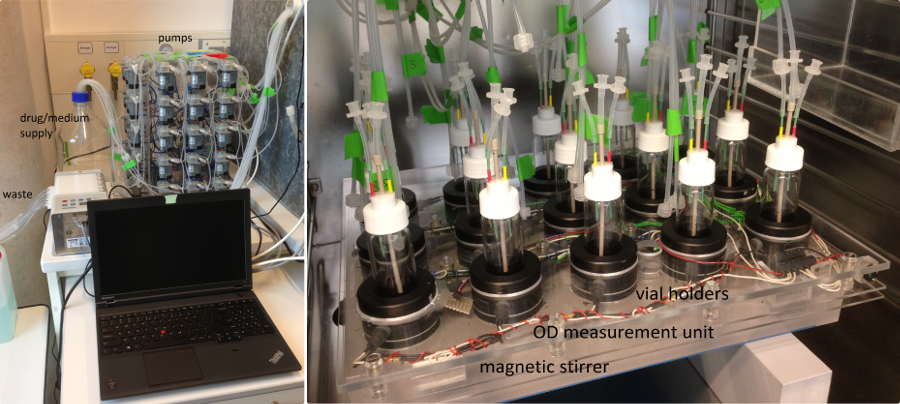 Our morbidostat set-up in Tübingen
Our morbidostat set-up in Tübingen
Analysis of bacterial pan-genomes
In addition to vertical inheritance, bacteria exchange plasmids and genomic DNA horizontally. This mix of inheritance patterns make the analysis of bacterial evolution challenging and interesting. To come to grips the complexity of bacterial genomes, Wei Ding has developed a pipe line to reconstruct and visualize bacterial pan-genomes. The output of this tool is available for exploration at pangenome.ch. We are currently exploring different ways to construct, visualize, and comprehend the evolutionary laws governing bacterial diversity.
Long-read sequencing and structural variation in bacterial genomes
Long-read sequencing technologies now allow cost-effective and large scale assembly of bacterial genomes. We are using Oxford Nanopore sequencing technology to characterize structural diversity in multi-drug resistant gram negative bacteria. Nicholas Noll has a first preprint on this work up on biorxiv. We developed a number of high-throughput criteria to assess the quality of fully assembled genomes for which curated references do not exist. Using this diverse collection of closed genomes and plasmids, we demonstrate rapid movement of carbapenemase between genomic neighborhoods, sequence types, and across species boundaries with distinct patterns for different carbapenemases. Ultimately, we hope to find new ways to track the spread of bacterial resistance determinants and thereby learn about the evolutionary dynamics of adaptation through horizontal transfer.
Publications
2025
Marco Molari, Liam P Shaw and Richard A Neher
Molecular Biology and Evolution, msae272. 10.1093/molbev/msae272 bibtex
2024
Julie Sollier, Marek Basler, Petr Broz
Nature Microbiology, 9 1--3. 10.1038/s41564-023-01566-w bibtex
2023
Liam P. Shaw and Richard A. Neher
Microbial Genomics, 9 001168. 10.1099/mgen.0.001168 bibtex
2023
Nicholas Noll, Marco Molari, Liam Shaw and Richard Neher
Microbial Genomics, in press., 2022.02.24.481757. 10.1101/2022.02.24.481757 bibtex
2019
Olivier Cunrath, Dominik M. Meinel, Pauline Maturana
EBioMedicine, 10.1016/j.ebiom.2019.02.061 bibtex
2018
Nicholas Noll, Eric Ulrich, Daniel Wuthrich
bioRxiv -- won't submit, 456897. 10.1101/456897 pdf bibtex
2018
Mumina Javed, Viola Ueltzhoeffer, Maximilian Heinrich
Journal of Antimicrobial Chemotherapy, 10.1093/jac/dky337 bibtex
2018
Talia L. Karasov, Juliana Almario, Claudia Friedemann
Cell Host & Microbe, 24 168--179.e4. 10.1016/j.chom.2018.06.011 pdf bibtex
2017
Wei Ding, Franz Baumdicker and Richard A. Neher
Nucleic Acids Research, 10.1093/nar/gkx977 pdf bibtex
2017
Bianca Doesselmann, Matthias Willmann, Matthias Steglich
Antimicrobial Agents and Chemotherapy, AAC.00043--17. 10.1128/AAC.00043-17 pdf bibtex