related posts
Human immunity is the main driving force of evolution of seasonal influenza viruses. Only when changing their antigenic properties, influenza viruses are able to re-infect previously infected humans. Mutation that prevent immune recognition often rapidly spread across the globe. For this reason, seasonal influenza vaccines need to be updated frequently. Vaccine strains are chosen among sampled virus strains, and the more closely this strain matches the future influenza virus population, the better the vaccine is going to be. Hence tracking influenza viruses is essential to maintain vaccine efficiency and accurate predictions of the composition of future populations could improve vaccines substantially.
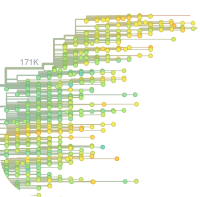
Real-time tracking of influenza virus evolution
Together with Trevor Bedford, we have been working on an interactive tool to explore influenza evolution. It is up and running at nextflu.org. In addition to a tree of the most recent influenza strains, users can explore the frequencies of mutations and clades of the tree. To facilitate data sharing and exploration during the recent ebola and MERS outbreaks, we have adapted nextflu and set up ebola.nextflu.org and mers.nextflu.org.
nextflu: Real-time tracking of seasonal influenza virus evolution in humans. Richard A. Neher, Trevor Bedford. Bioinformatics, btv381
Predicting influenza evolution
In collaboration with Colin Russell and Boris Shraiman — we have shown that it is possible to predict which individual from a population is most closely related to future populations. The method uses the branching pattern of genealogical trees to estimate which part of the tree contains the “fittest” sequences. When applied to historical influenza data, the method makes informative predictions in about 80% of the time.
Enterovirus D-68
Worldwide outbreaks of enterovirus D68 (EV-D68) in 2014 and 2016 have caused widespread serious respiratory and neurological disease. In collaboration with Jan Albert and Robert Dyrdak in Stockholm, Emma Hodcroft is investigating the spread and diversity of this virus using whole genome deep sequencing. Phylogenetic trees reconstructed from the sequences suggest that EV-D68 was introduced into Stockholm several times during the 2016 outbreak and then spread locally. Putative neutralization targets in the BC- and DE-loops of the VP1 protein are slightly more diverse within-host and tend to undergo more frequent substitution than other genomic regions. The new data along with publicly available EV-D68 sequences are included in an interactive phylodynamic analysis on nextstrain to facilitate timely EV-D68 tracking in the future.
Publications
2017
Dylan H. Morris, Katelyn M. Gostic, Simone Pompei
Trends in Microbiology, 10.1016/j.tim.2017.09.004 bibtex
2016
Richard A. Neher, Trevor Bedford, Rodney S. Daniels, Colin A. Russell and Boris I. Shraiman
Proceedings of the National Academy of Sciences of the United States of America, 113 E1701--1709. 10.1073/pnas.1525578113 pdf bibtex
2015
Richard A. Neher and Trevor Bedford
Bioinformatics (Oxford, England), 31 3546--3548. 10.1093/bioinformatics/btv381 pdf bibtex
2014
Richard A. Neher, Colin A. Russell and Boris I. Shraiman
eLife, 3 10.7554/eLife.03568 pdf bibtex